Survey Data with Pandas and StatsModels#
Tutorial for PyCon 2025
Allen B. Downey
Click here to run this notebook on Colab.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
Show code cell content
# Configure Matplotlib
plt.rcParams["figure.figsize"] = 7, 3.5
plt.rcParams["figure.dpi"] = 75
plt.rcParams["axes.titlelocation"] = "left"
plt.rcParams["axes.spines.top"] = False
plt.rcParams["axes.spines.bottom"] = False
plt.rcParams["axes.spines.left"] = False
plt.rcParams["axes.spines.right"] = False
from os.path import basename, exists
def download(url):
filename = basename(url)
if not exists(filename):
from urllib.request import urlretrieve
local, _ = urlretrieve(url, filename)
print('Downloaded ' + local)
def decorate(**options):
"""Decorate the current axes.
Call decorate with keyword arguments like
decorate(title='Title',
xlabel='x',
ylabel='y')
The keyword arguments can be any of the axis properties
https://matplotlib.org/api/axes_api.html
"""
legend = options.pop("legend", True)
loc = options.pop("loc", "best")
# Pass options to Axis.set
ax = plt.gca()
ax.set(**options)
# Add a legend if there are any labeled elements
handles, labels = ax.get_legend_handles_labels()
if handles and legend:
ax.legend(handles, labels, loc=loc)
# Tight layout is generally a good idea
plt.tight_layout()
Data#
We will use data from the General Social Survey (GSS). The raw dataset is big, so I’ve prepared an extract, which the following cell downloads. The code to generate the extract is in this notebook.
# This dataset is prepared in the GssExtract repository
DATA_PATH = "https://github.com/AllenDowney/GssExtract/raw/main/data/interim/"
filename = "gss_extract_pacs_workshop.hdf"
download(DATA_PATH + filename)
Show code cell content
# Solution
gss = pd.read_hdf(filename, "gss")
gss.shape
(72390, 29)
Show code cell content
# Solution
gss.head()
| age | attend | ballot | class | cohort | degree | educ | fair | fear | goodlife | ... | region | relig | rincome | satfin | satjob | sex | srcbelt | trust | wtssall | year | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 24.0 | 1.0 | NaN | 3.0 | 1948.0 | 3.0 | 16.0 | 2.0 | NaN | NaN | ... | 1 | 1.0 | NaN | 1.0 | NaN | 1.0 | 5.0 | 1.0 | 1.3339 | 1972 |
| 1 | 33.0 | 2.0 | NaN | 2.0 | 1939.0 | 0.0 | 10.0 | 2.0 | NaN | NaN | ... | 1 | 2.0 | NaN | 3.0 | 2.0 | 1.0 | 1.0 | 1.0 | 0.8893 | 1972 |
| 2 | 40.0 | 8.0 | NaN | 2.0 | 1932.0 | 1.0 | 12.0 | 2.0 | NaN | NaN | ... | 1 | 5.0 | NaN | 1.0 | NaN | 2.0 | 3.0 | 2.0 | 1.3339 | 1972 |
| 3 | 24.0 | 2.0 | NaN | 2.0 | 1948.0 | 1.0 | 12.0 | 1.0 | NaN | NaN | ... | 9 | 2.0 | NaN | 2.0 | NaN | 1.0 | 3.0 | 2.0 | 1.7786 | 1972 |
| 4 | 21.0 | 7.0 | NaN | 3.0 | 1951.0 | 1.0 | 12.0 | 1.0 | NaN | NaN | ... | 9 | 1.0 | NaN | 1.0 | NaN | 2.0 | 2.0 | NaN | 1.7786 | 1972 |
5 rows × 29 columns
Show code cell content
# Solution
gss.columns
Index(['age', 'attend', 'ballot', 'class', 'cohort', 'degree', 'educ', 'fair',
'fear', 'goodlife', 'hapmar', 'happy', 'health', 'helpful', 'id',
'life', 'partyid', 'polviews', 'race', 'region', 'relig', 'rincome',
'satfin', 'satjob', 'sex', 'srcbelt', 'trust', 'wtssall', 'year'],
dtype='object')
Show code cell content
# Solution
gss.isna().mean().sort_values()
id 0.000000
year 0.000000
wtssall 0.000000
region 0.000000
srcbelt 0.000718
race 0.001796
sex 0.002086
degree 0.002942
educ 0.004338
cohort 0.005567
relig 0.007087
partyid 0.007294
attend 0.009711
age 0.010416
class 0.048501
satfin 0.063641
happy 0.065106
polviews 0.131620
health 0.236552
satjob 0.265064
ballot 0.302183
fear 0.365147
life 0.386669
rincome 0.402086
trust 0.410982
helpful 0.414090
fair 0.415665
hapmar 0.468241
goodlife 0.631924
dtype: float64
Distributions#
We’ll see two ways to describe distributions: value counts and KDE.
And for joint distributions, we’ll use a cross tabulation.
def value_counts(seq, **options):
"""Make a series of values and the number of times they appear.
Args:
seq: sequence
returns: Pandas Series
"""
return pd.Series(seq).value_counts(**options).sort_index()
Show code cell content
# Solution
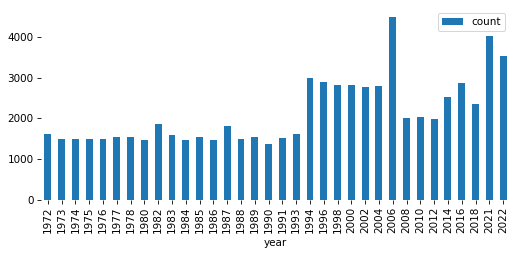
pmf = value_counts(gss['year'])
pmf.plot(kind='bar')
decorate()
Show code cell content
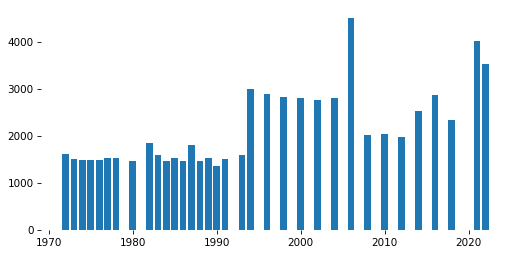
# Solution
x, y = pmf.index, pmf.values
plt.bar(x, y)
decorate()
Show code cell content
# Solution
gss['age'].describe()
count 71636.000000
mean 44.985943
std 17.201666
min 18.000000
25% 30.000000
50% 43.000000
75% 58.000000
max 89.000000
Name: age, dtype: float64
Show code cell content
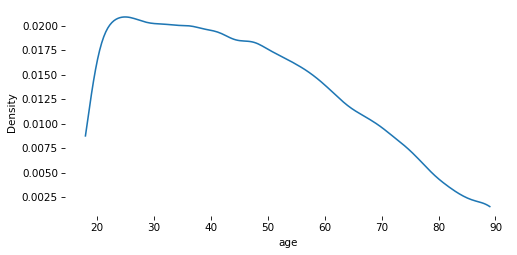
# Solution
sns.kdeplot(gss['age'], cut=0)
decorate()
Show code cell content
# Solution
gss['cohort'].describe()
count 71987.000000
mean 1991.908095
std 561.717010
min 1883.000000
25% 1938.000000
50% 1954.000000
75% 1968.000000
max 9999.000000
Name: cohort, dtype: float64
Show code cell content
# Solution
gss['cohort'] = gss['cohort'].replace(9999, np.nan)
Exercise#
Use value_counts to plot the distribution of cohort as a bar plot.
Then use sns.kdeplot to show the same distribution as a continuous quantity.
Show code cell content
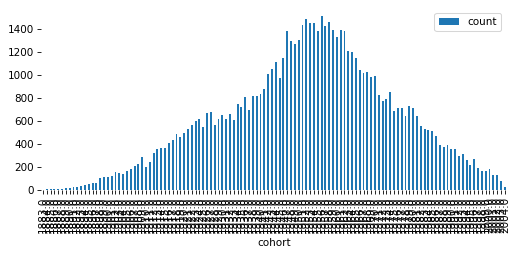
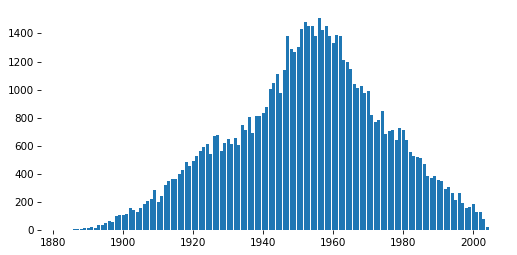
# Solution
pmf = value_counts(gss['cohort'])
pmf.plot(kind='bar')
decorate(ylabel='Count')
Show code cell content
# Solution
x, y = pmf.index, pmf.values
plt.bar(x, y)
decorate(ylabel='Count')
Show code cell content
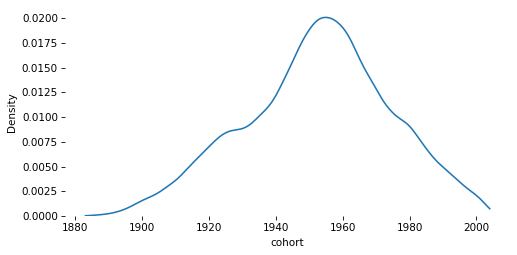
# Solution
sns.kdeplot(gss['cohort'], cut=0)
decorate()
Cross tabulation#
Cross tabulation is a way to represent the joint distribution of two variables.
Show code cell content
# Solution
xtab = pd.crosstab(gss['cohort'], gss['age'])
xtab
| age | 18.0 | 19.0 | 20.0 | 21.0 | 22.0 | 23.0 | 24.0 | 25.0 | 26.0 | 27.0 | ... | 80.0 | 81.0 | 82.0 | 83.0 | 84.0 | 85.0 | 86.0 | 87.0 | 88.0 | 89.0 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cohort | |||||||||||||||||||||
| 1883.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1884.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 1885.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| 1886.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 |
| 1887.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2000.0 | 36 | 0 | 0 | 71 | 77 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2001.0 | 0 | 0 | 68 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2002.0 | 0 | 59 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2003.0 | 7 | 70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2004.0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
122 rows × 72 columns
Show code cell content
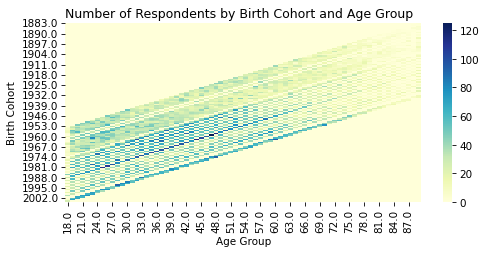
# Solution
sns.heatmap(
xtab,
cmap="YlGnBu",
)
plt.title("Number of Respondents by Birth Cohort and Age Group")
plt.xlabel("Age Group")
plt.ylabel("Birth Cohort")
decorate()
Target variable#
I’ve selected four variables you might want to work with. Whichever one you choose, put its cell last.
# https://gssdataexplorer.norc.org/variables/434/vshow
varname = 'happy'
question = """Taken all together, how would you say things are these days--
would you say that you are very happy, pretty happy, or not too happy?
"""
responses = ['Very happy', "Happy", 'Not too happy']
ylabel = "Percent saying 'very happy'"
# https://gssdataexplorer.norc.org/variables/439/vshow
varname = 'helpful'
question = """Would you say that most of the time people try to be helpful,
or that they are mostly just looking out for themselves?
"""
responses = ['Helpful', 'Look out\nfor themselves', 'Depends']
ylabel = "Percent saying 'helpful'"
# https://gssdataexplorer.norc.org/variables/440/vshow
varname = 'fair'
question = """Do you think most people would try to take advantage of you
if they got a chance, or would they try to be fair?
"""
# notice that the negative response is first here!
responses = ['Take advantage', 'Fair', 'Depends']
ylabel = "Percent saying 'fair'"
# https://gssdataexplorer.norc.org/variables/441/vshow
varname = 'trust'
question = """Generally speaking, would you say that most people can be trusted
or that you can't be too careful in dealing with people?
"""
responses = ['Trusted', "Can't be\ntoo careful", 'Depends']
ylabel = "Percent saying 'trusted'"
Show code cell content
# Solution
column = gss[varname]
column
0 1.0
1 1.0
2 2.0
3 2.0
4 NaN
...
72385 2.0
72386 2.0
72387 NaN
72388 NaN
72389 1.0
Name: trust, Length: 72390, dtype: float64
Show code cell content
# Solution
value_counts(column, dropna=False)
trust
1.0 15783
2.0 24890
3.0 1966
NaN 29751
Name: count, dtype: int64
Show code cell content
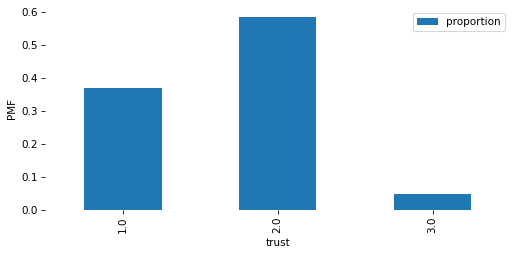
# Solution
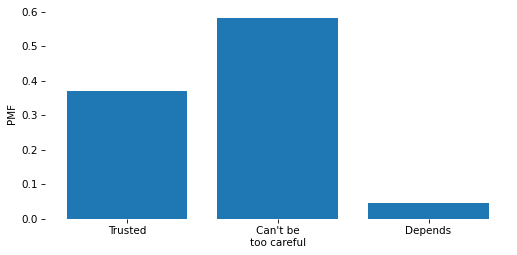
pmf = value_counts(column, normalize=True)
pmf
trust
1.0 0.370154
2.0 0.583738
3.0 0.046108
Name: proportion, dtype: float64
Show code cell content
# Solution
pmf.plot(kind='bar')
decorate(ylabel='PMF')
Show code cell content
# Solution
plt.bar(pmf.index, pmf)
plt.xticks(pmf.index, responses)
decorate(ylabel='PMF')
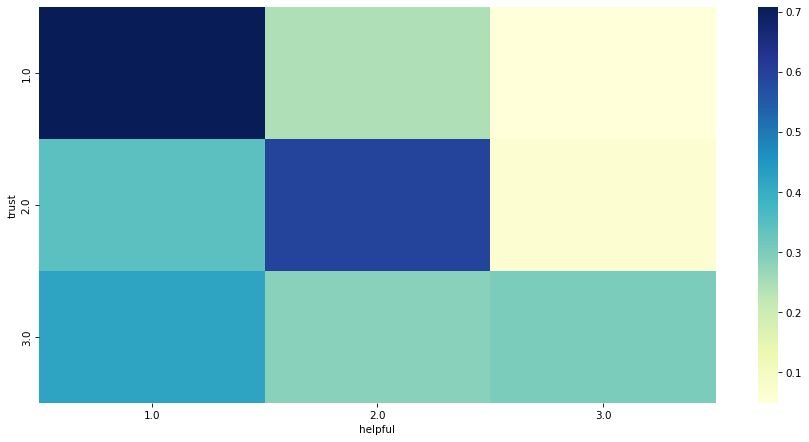
Exercise#
Do you think people who say “trust” are also more likely to say “helpful”?
To find out, make a cross tabulation of trust and helpful.
Consider using normalize='index'.
Make a heatmap that shows this joint distribution.
What relationships can you see in the responses?
Show code cell content
# Solution
xtab = pd.crosstab(gss['trust'], gss['helpful'], normalize='index')
xtab
| helpful | 1.0 | 2.0 | 3.0 |
|---|---|---|---|
| trust | |||
| 1.0 | 0.707773 | 0.243133 | 0.049094 |
| 2.0 | 0.340947 | 0.592560 | 0.066493 |
| 3.0 | 0.417197 | 0.283970 | 0.298832 |
Show code cell content
# Solution
plt.figure(figsize=(12, 6))
sns.heatmap(
xtab,
cmap="YlGnBu",
)
decorate(xlabel='helpful', ylabel='trust')
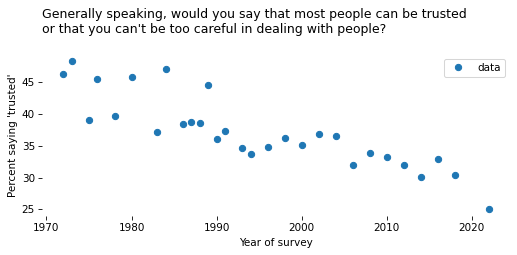
Time Series#
To see how responses have changed over time, we’ll use cross tabulation again.
Show code cell content
# Solution
xtab = pd.crosstab(gss["year"], gss[varname], normalize='index')
xtab.head()
| trust | 1.0 | 2.0 | 3.0 |
|---|---|---|---|
| year | |||
| 1972 | 0.463551 | 0.490343 | 0.046106 |
| 1973 | 0.483656 | 0.495664 | 0.020680 |
| 1975 | 0.391099 | 0.564396 | 0.044504 |
| 1976 | 0.455518 | 0.513043 | 0.031438 |
| 1978 | 0.396597 | 0.549738 | 0.053665 |
Show code cell content
# Solution
gss72 = gss.query('year == 1972')
gss72.shape
(1613, 29)
Show code cell content
# Solution
pmf72 = value_counts(gss72[varname], normalize=True)
pmf72
trust
1.0 0.463551
2.0 0.490343
3.0 0.046106
Name: proportion, dtype: float64
Show code cell content
# Solution
time_series = xtab[1] * 100
time_series.name = "data"
Show code cell content
# Solution
time_series.plot(style='o')
decorate(
xlabel="Year of survey",
ylabel=ylabel,
title=question,
)
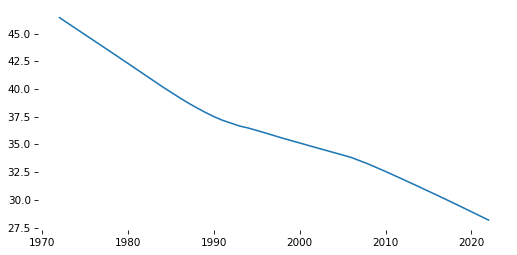
Smoothing#
To smooth the time series, we’ll use LOWESS (locally weighted scatterplot smoothing).
Show code cell content
# Solution
x = time_series.index.to_numpy()
y = time_series.to_numpy()
Show code cell content
# Solution
from statsmodels.nonparametric.smoothers_lowess import lowess
smooth_array = lowess(y, x)
Show code cell content
# Solution
index, data = np.transpose(smooth_array)
smooth_series = pd.Series(data, index=index)
Show code cell content
# Solution
smooth_series.plot()
decorate()
def make_lowess(series, frac=0.5):
"""Use LOWESS to compute a smooth line.
series: pd.Series
returns: pd.Series
"""
y = series.to_numpy()
x = series.index.to_numpy()
smooth_array = lowess(y, x, frac)
index, data = np.transpose(smooth_array)
return pd.Series(data, index=index)
Exercise#
Use this function to make smooth_series.
Plot the result along with time_series.
Show code cell content
# Solution
smooth_series = make_lowess(time_series)
Show code cell content
# Solution
time_series.plot(style='o', alpha=0.5)
smooth_series.plot(label='smoothed', color='C0')
decorate(
xlabel="Year of survey",
ylabel=ylabel,
title=question,
)
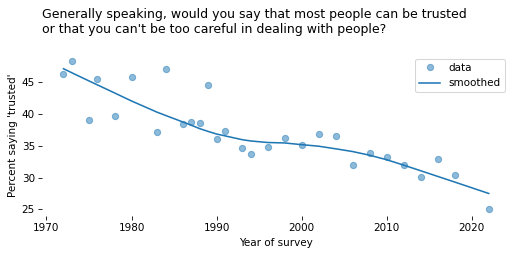
Binary series#
It will be useful to convert the responses to a binary variable represented with 0s and 1s.
Show code cell content
# Solution
# tempting but wrong
gss['y'] = gss[varname] == 1
value_counts(gss['y'], dropna=False)
y
False 56607
True 15783
Name: count, dtype: int64
Show code cell content
# Solution
gss['y'] = np.where(gss[varname].notna(), gss[varname] == 1, np.nan)
value_counts(gss['y'], dropna=False)
y
0.0 26856
1.0 15783
NaN 29751
Name: count, dtype: int64
Show code cell content
# Solution
# Reminder: this needs to be updated depending on the target variable
gss['y'] = gss[varname].replace([1, 2, 3], [1, 0, 0])
value_counts(gss['y'], dropna=False)
y
0.0 26856
1.0 15783
NaN 29751
Name: count, dtype: int64
Pivot tables#
Now we’ll use a pivot table to put respondents in groups and see how the responses have changed over time.
polviews_map = {
1: 'Liberal',
2: 'Liberal',
3: 'Liberal',
4: 'Moderate',
5: 'Conservative',
6: 'Conservative',
7: 'Conservative',
}
Show code cell content
# Solution
gss['polviews3'] = gss['polviews'].replace(polviews_map)
Show code cell content
# Solution
value_counts(gss['polviews3'], dropna=False)
polviews3
Conservative 21580
Liberal 17370
Moderate 23912
NaN 9528
Name: count, dtype: int64
Show code cell content
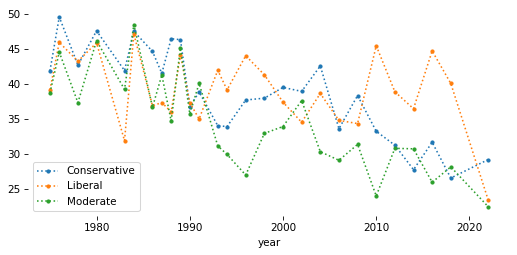
# Solution
table = gss.pivot_table(
index='year', columns='polviews3', values='y', aggfunc='mean'
) * 100
table.head()
| polviews3 | Conservative | Liberal | Moderate |
|---|---|---|---|
| year | |||
| 1975 | 41.891892 | 39.192399 | 38.733706 |
| 1976 | 49.590164 | 45.961003 | 44.561404 |
| 1978 | 42.699115 | 43.262411 | 37.347295 |
| 1980 | 47.609562 | 45.896657 | 46.204620 |
| 1983 | 41.935484 | 31.914894 | 39.240506 |
Show code cell content
# Solution
table.plot(style='.:')
decorate()
Better colors#
We’ll select colors from a Seaborn color palette.
Show code cell content
# Solution
muted = sns.color_palette('muted', 5)
sns.palplot(muted)
Show code cell content
# Solution
color_map = {
'Conservative': muted[3],
'Moderate': muted[4],
'Liberal': muted[0]
}
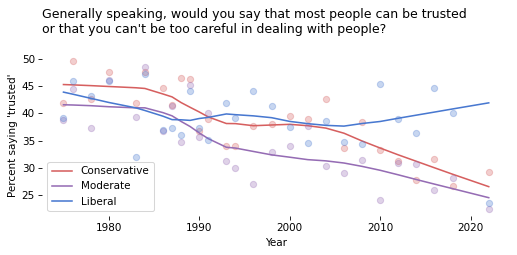
Exercise#
Loop through the groups in color_map.
For each one, extract a column from table and plot the data points; then plot a smooth line.
Show code cell content
# Solution
for group in color_map:
series = table[group]
color = color_map[group]
series.plot(style='o', label='', color=color, alpha=0.3)
smooth = make_lowess(series)
smooth.plot(label=group, color=color)
decorate(
xlabel='Year',
ylabel=ylabel,
title=question,
)
Groupby#
Now that we have y as a binary variable, we have another way to compute the time series and the pivot table.
There are pros and cons of each method.
Show code cell content
# Solution
valid = gss.dropna(subset=['y'])
Show code cell content
# Solution
time_series2 = valid.groupby('year')['y'].mean() * 100
assert np.allclose(time_series, time_series2)
Show code cell content
# Solution
table2 = valid.groupby(['year', 'polviews3'])['y'].mean().unstack() * 100
assert np.allclose(table, table2)
Logistic regression#
We’ll use logistic regression to model changes over time.
Show code cell content
# Solution
year_shift = gss['year'].median()
gss['x'] = gss['year'] - year_shift
Show code cell content
# Solution
subset = gss.dropna(subset=['y', 'x'])
Show code cell content
# Solution
import statsmodels.formula.api as smf
formula = 'y ~ x + I(x**2)'
model = smf.logit(formula, data=subset).fit(disp=False)
model.params
Intercept -0.588941
x -0.014565
I(x ** 2) 0.000057
dtype: float64
Show code cell content
# Solution
year_range = np.arange(1972, 2026)
pred_df = pd.DataFrame(dict(x=year_range - year_shift))
pred_series = model.predict(pred_df) * 100
pred_series.index = year_range
Show code cell content
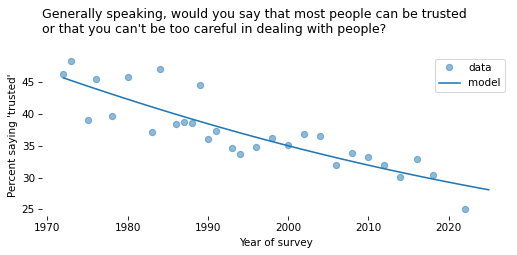
# Solution
time_series.plot(style='o', alpha=0.5)
pred_series.plot(label='model', color='C0')
decorate(
xlabel="Year of survey",
ylabel=ylabel,
title=question,
)
def fit_model(data, x_range, x_shift):
formula = 'y ~ x + I(x**2)'
model = smf.logit(formula, data=data).fit(disp=False)
pred_df = pd.DataFrame(dict(x=x_range - x_shift))
pred = model.predict(pred_df) * 100
pred.index = x_range
pred.name = 'model'
return pred
Show code cell content
# Solution
pred_series2 = fit_model(subset, year_range, year_shift)
assert np.allclose(pred_series, pred_series2)
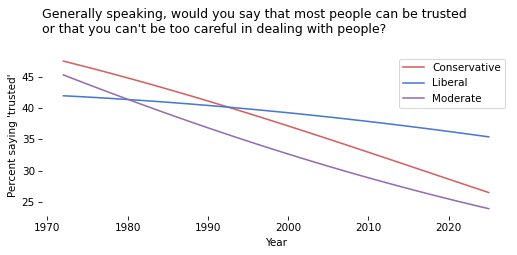
Exercise#
Use subset.groupby('polviews3') to loop through the groups.
For each one, use fit_model to generate and plot a “prediction” for the group.
Show code cell content
# Solution
for name, group in subset.groupby('polviews3'):
pred_series = fit_model(group, year_range, year_shift)
pred_series.plot(label=name, color=color_map[name])
decorate(
xlabel='Year',
ylabel=ylabel,
title=question,
)
Group by cohort#
Now let’s group respondents by decade of birth and see how each cohort changes over time.
def round_into_bins(series, bin_width, low=0, high=None):
"""Rounds values down to the bin they belong in.
series: pd.Series
bin_width: number, width of the bins
returns: Series of bin values (with NaN preserved)
"""
if high is None:
high = series.max()
bins = np.arange(low, high + bin_width, bin_width)
indices = np.digitize(series, bins)
result = pd.Series(bins[indices - 1], index=series.index, dtype='float')
result[series.isna()] = np.nan
return result
Show code cell content
# Solution
gss["cohort10"] = round_into_bins(gss["cohort"], 10, 1880)
value_counts(gss["cohort10"], dropna=False)
cohort10
1880.0 45
1890.0 501
1900.0 1722
1910.0 3616
1920.0 5862
1930.0 7115
1940.0 10925
1950.0 14273
1960.0 11699
1970.0 7681
1980.0 5088
1990.0 2563
2000.0 545
NaN 755
Name: count, dtype: int64
Show code cell content
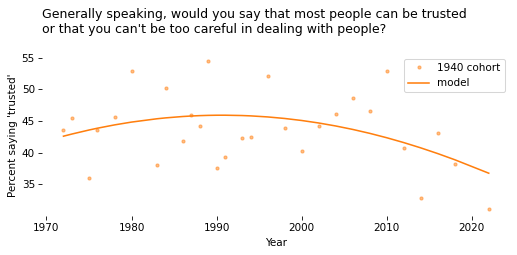
# Solution
cohort_df = gss.query("cohort10 == 1940").dropna(subset=['y', 'x'])
cohort_df.shape
(6825, 33)
Show code cell content
# Solution
cohort_series = cohort_df.groupby('year')['y'].mean() * 100
cohort_series.name = '1940 cohort'
Show code cell content
# Solution
x_range = cohort_series.index
pred_series = fit_model(cohort_df, x_range, year_shift)
Show code cell content
# Solution
cohort_series.plot(style='.', color='C1', alpha=0.5)
pred_series.plot(color='C1')
decorate(
xlabel="Year",
ylabel=ylabel,
title=question,
)
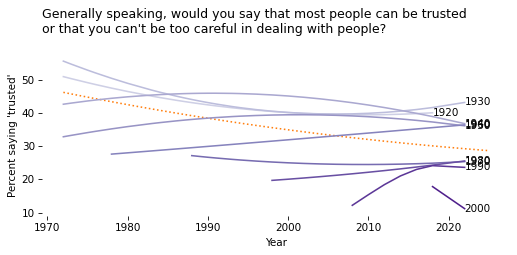
All the Cohorts#
Now let’s compare cohorts. The following function encapsulates the steps from the previous section.
def plot_cohort(df, cohort, color='C0'):
cohort_df = df.query("cohort10 == @cohort").dropna(subset=['y', 'x'])
cohort_series = cohort_df.groupby('year')['y'].mean() * 100
year_range = cohort_series.index
pred_series = fit_model(cohort_df, year_range, year_shift)
x, y = pred_series.index[-1], pred_series.values[-1]
plt.text(x, y, cohort, ha='left', va='center')
pred_series.plot(label=cohort, color=color)
Show code cell content
# Solution
first, last = 1920, 2000
subset = gss.query("@first <= cohort <= @last" )
subset.shape
(65390, 33)
Show code cell content
# Solution
pred_all = fit_model(subset, year_range, year_shift)
Show code cell content
# Solution
cohorts = np.sort(subset['cohort10'].unique().astype(int))
cohorts
array([1920, 1930, 1940, 1950, 1960, 1970, 1980, 1990, 2000])
Show code cell content
# Solution
cmap = plt.get_cmap('Purples')
colors = [cmap(x) for x in np.linspace(0.3, 0.9, len(cohorts))]
Exercise#
Plot pred_all.
Then loop through cohorts and colors, and call plot_cohort for each group.
Depending on which target variable you chose, you might get a warning from StatsModels.
Show code cell content
# Solution
pred_all.plot(ls=':', color='C1')
for cohort, color in zip(cohorts, colors):
plot_cohort(gss, cohort, color)
decorate(
xlabel="Year",
ylabel=ylabel,
title=question,
legend=False
)
/home/downey/miniconda3/envs/SurveyDataPandas/lib/python3.13/site-packages/statsmodels/base/model.py:607: ConvergenceWarning: Maximum Likelihood optimization failed to converge. Check mle_retvals
warnings.warn("Maximum Likelihood optimization failed to "
Simpson’s Paradox#
The composition of the population changes over time.
Show code cell content
# Solution
xtab_cohort = pd.crosstab(gss["cohort10"], gss["year"], normalize="columns")
xtab_cohort.head()
| year | 1972 | 1973 | 1974 | 1975 | 1976 | 1977 | 1978 | 1980 | 1982 | 1983 | ... | 2004 | 2006 | 2008 | 2010 | 2012 | 2014 | 2016 | 2018 | 2021 | 2022 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cohort10 | |||||||||||||||||||||
| 1880.0 | 0.003113 | 0.003995 | 0.008108 | 0.006739 | 0.004692 | 0.000000 | 0.003277 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 |
| 1890.0 | 0.047945 | 0.038615 | 0.029730 | 0.037062 | 0.047587 | 0.027613 | 0.028834 | 0.017088 | 0.018929 | 0.005013 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 |
| 1900.0 | 0.110212 | 0.099201 | 0.129730 | 0.082210 | 0.070375 | 0.091387 | 0.083224 | 0.059467 | 0.051379 | 0.038221 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 |
| 1910.0 | 0.152553 | 0.158455 | 0.131081 | 0.130728 | 0.144772 | 0.125575 | 0.105505 | 0.124402 | 0.103840 | 0.093358 | ... | 0.015714 | 0.007562 | 0.007944 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 |
| 1920.0 | 0.224782 | 0.180426 | 0.193243 | 0.187332 | 0.159517 | 0.184747 | 0.152687 | 0.160629 | 0.149270 | 0.137218 | ... | 0.038929 | 0.040036 | 0.038232 | 0.035294 | 0.025381 | 0.014235 | 0.009807 | 0.008576 | 0.0 | 0.0 |
5 rows × 34 columns
Show code cell content
# Solution
xtab_cohort[1972].plot(label="1972")
xtab_cohort[1998].plot(label="1998")
xtab_cohort[2022].plot(label="2022")
decorate(xlabel="Year of birth", ylabel="Fraction of sample")
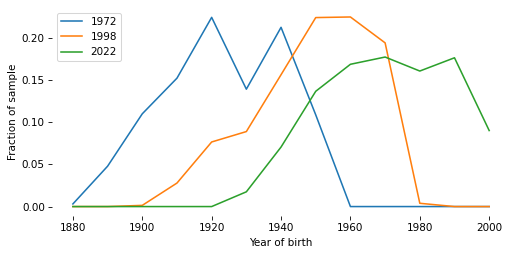
Sampling weights#
The GSS uses stratified sampling, so some respondents represent more people in the population than others. We can use resampling to correct for stratified sampling.
Show code cell content
# Solution
gss['wtssall'].describe()
count 72390.000000
mean 1.303960
std 0.889990
min 0.113721
25% 0.918400
50% 1.062700
75% 1.547800
max 14.272462
Name: wtssall, dtype: float64
Show code cell content
# Solution
gss.groupby('year')['wtssall'].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| year | ||||||||
| 1972 | 1613.0 | 1.146481 | 0.486191 | 0.444600 | 0.889300 | 0.889300 | 1.333900 | 3.557100 |
| 1973 | 1504.0 | 1.130645 | 0.417802 | 0.457200 | 0.914500 | 0.914500 | 1.371700 | 2.743500 |
| 1974 | 1484.0 | 1.160032 | 0.443681 | 0.466000 | 0.932000 | 0.932000 | 1.397900 | 3.727900 |
| 1975 | 1490.0 | 1.140258 | 0.466167 | 0.466000 | 0.932000 | 0.932000 | 1.397900 | 3.261900 |
| 1976 | 1499.0 | 1.147392 | 0.459938 | 0.485400 | 0.970900 | 0.970900 | 1.456300 | 3.398100 |
| 1977 | 1530.0 | 1.161231 | 0.458057 | 0.494100 | 0.988100 | 0.988100 | 1.482200 | 2.964400 |
| 1978 | 1532.0 | 1.148797 | 0.448502 | 0.506300 | 1.012700 | 1.012700 | 1.519000 | 3.038000 |
| 1980 | 1468.0 | 1.168663 | 0.486058 | 0.511200 | 1.022500 | 1.022500 | 1.533700 | 3.578700 |
| 1982 | 1860.0 | 1.189170 | 0.513264 | 0.506400 | 1.031900 | 1.031900 | 1.547800 | 4.127500 |
| 1983 | 1599.0 | 1.174535 | 0.489286 | 0.505800 | 1.011600 | 1.011600 | 1.517500 | 3.540700 |
| 1984 | 1473.0 | 1.163876 | 0.473032 | 0.516500 | 1.033100 | 1.033100 | 1.549600 | 3.615700 |
| 1985 | 1534.0 | 1.158889 | 0.463941 | 0.518100 | 1.036300 | 1.036300 | 1.554400 | 3.626900 |
| 1986 | 1470.0 | 1.164447 | 0.467082 | 0.503600 | 1.007200 | 1.007200 | 1.510800 | 3.021600 |
| 1987 | 1819.0 | 1.186371 | 0.499286 | 0.505200 | 1.010400 | 1.010400 | 1.515500 | 3.536200 |
| 1988 | 1481.0 | 1.163860 | 0.463531 | 0.531300 | 1.062700 | 1.062700 | 1.062700 | 3.719400 |
| 1989 | 1537.0 | 1.184300 | 0.504330 | 0.510700 | 1.021500 | 1.021500 | 1.532200 | 3.575100 |
| 1990 | 1372.0 | 1.161439 | 0.489095 | 0.531300 | 1.062700 | 1.062700 | 1.062700 | 3.188100 |
| 1991 | 1517.0 | 1.174147 | 0.478842 | 0.531100 | 1.062100 | 1.062100 | 1.593200 | 3.717500 |
| 1993 | 1606.0 | 1.169310 | 0.448682 | 0.528500 | 1.057100 | 1.057100 | 1.585600 | 3.699800 |
| 1994 | 2992.0 | 1.166058 | 0.457253 | 0.541200 | 1.082500 | 1.082500 | 1.082500 | 3.247500 |
| 1996 | 2904.0 | 1.181024 | 0.479229 | 0.543200 | 1.086400 | 1.086400 | 1.086400 | 3.259100 |
| 1998 | 2832.0 | 1.177809 | 0.462729 | 0.550100 | 1.100100 | 1.100100 | 1.100100 | 3.850400 |
| 2000 | 2817.0 | 1.165950 | 0.453726 | 0.549300 | 1.098500 | 1.098500 | 1.098500 | 3.295600 |
| 2002 | 2765.0 | 1.198505 | 0.507470 | 0.557800 | 1.115600 | 1.115600 | 1.115600 | 2.788900 |
| 2004 | 2812.0 | 1.299424 | 0.762699 | 0.459200 | 0.918400 | 0.961700 | 1.836800 | 6.428700 |
| 2006 | 4510.0 | 1.360152 | 0.841060 | 0.429700 | 0.859300 | 0.954800 | 1.909600 | 5.728800 |
| 2008 | 2023.0 | 1.425750 | 0.911863 | 0.437745 | 0.875490 | 1.067670 | 2.135340 | 6.406021 |
| 2010 | 2044.0 | 1.313222 | 0.726111 | 0.463128 | 0.926255 | 0.926255 | 1.684101 | 4.210252 |
| 2012 | 1974.0 | 1.366495 | 0.884637 | 0.411898 | 0.823796 | 1.235694 | 1.747975 | 8.739876 |
| 2014 | 2538.0 | 1.273430 | 0.674193 | 0.448002 | 0.896003 | 1.344005 | 1.380591 | 5.376020 |
| 2016 | 2867.0 | 1.227412 | 0.607020 | 0.391825 | 0.956994 | 0.956994 | 1.564363 | 4.306471 |
| 2018 | 2348.0 | 1.382192 | 0.936160 | 0.471499 | 0.942997 | 0.942997 | 1.414496 | 5.897420 |
| 2021 | 4032.0 | 1.551637 | 1.175611 | 0.227357 | 0.790829 | 1.195842 | 1.869697 | 7.557038 |
| 2022 | 3544.0 | 2.551821 | 2.489079 | 0.113721 | 0.714314 | 1.781121 | 3.529911 | 14.272462 |
def resample_by_year(df, column='wtssall'):
"""Resample rows within each year using weighted sampling.
df: DataFrame
column: string name of weight variable
returns: DataFrame
"""
grouped = df.groupby('year')
samples = [group.sample(n=len(group), replace=True, weights=group[column])
for _, group in grouped]
sample = pd.concat(samples, ignore_index=True)
return sample
Show code cell content
# Solution
sample = resample_by_year(gss)
Show code cell content
# Solution
sample['y'].mean()
np.float64(0.36038778568752344)
Show code cell content
# Solution
gss['y'].mean()
np.float64(0.3701540842890312)
Show code cell content
# Solution
weighted_mean = np.average(valid['y'], weights=valid['wtssall'])
Copyright 2025 Allen Downey
The code in this notebook is under the MIT license.