Power law? Or just a guideline?#
Part two. See ctokens.ipynb for part one.
# Install empiricaldist if we don't already have it
try:
import empiricaldist
except ImportError:
!pip install empiricaldist
from os.path import basename, exists
def download(url):
filename = basename(url)
if not exists(filename):
from urllib.request import urlretrieve
local, _ = urlretrieve(url, filename)
print("Downloaded " + local)
download(
"https://github.com/AllenDowney/ProbablyOverthinkingIt/raw/book/notebooks/utils.py"
)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from utils import decorate
from utils import empirical_error_bounds
from utils import plot_error_bounds
# Make the figures smaller to save some screen real estate
plt.rcParams["figure.dpi"] = 75
plt.rcParams["figure.figsize"] = [6, 3.5]
plt.rcParams["axes.titlelocation"] = "left"
Data#
First, let’s download the data and supporting scripts from Hatton’s site.
download(
"http://www.leshatton.org/Documents/HattonWarr_SharedProps_Apr2025.zip"
)
Here are the files in the zip archive.
import zipfile
zip_path = "HattonWarr_SharedProps_Apr2025.zip"
with zipfile.ZipFile(zip_path, 'r') as zf:
file_list = zf.namelist()
Here’s the script that generates the PMF of token lengths.
target_file = "3-fig4AB.sh"
with zipfile.ZipFile(zip_path, 'r') as zf:
with zf.open(target_file) as f:
# Read as bytes and decode to string
contents = f.read().decode("utf-8")
Now we can put the results in a DataFrame.
target_file = "24-06_trembl_pdf.dat"
with zipfile.ZipFile(zip_path, "r") as zf:
with zf.open(target_file) as f:
df = pd.read_csv(
f,
comment="#", # skip header lines starting with '#'
delim_whitespace=True,
names=["value", "freq"]
)
Before we do anything else, let’s compute the log of the token lengths.
df['mag'] = np.log10(df['value'])
df.head()
| value | freq | mag | |
|---|---|---|---|
| 0 | 7 | 130 | 0.845098 |
| 1 | 8 | 10885 | 0.903090 |
| 2 | 9 | 16432 | 0.954243 |
| 3 | 10 | 17303 | 1.000000 |
| 4 | 11 | 14607 | 1.041393 |
And the total number of values.
count = df['freq'].sum()
count
251254507
Here’s a Pmf of the magnitudes.
from empiricaldist import Pmf
pmf = Pmf(df['freq'].values, df['mag'])
pmf.normalize()
251254507
The following function computes the tail distribution (fraction of values >= x for all x).
from empiricaldist import Pmf, Surv
def make_surv(pmf):
"""Make a non-standard survival function, P(X>=x)"""
surv = pmf.make_surv() + pmf
# correct for numerical error
surv.iloc[0] = 1
return Surv(surv)
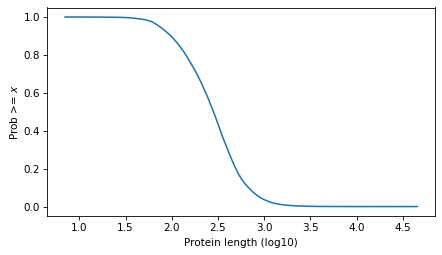
Here’s what it looks like.
surv = make_surv(pmf)
xlabel = "Protein length (log10)"
ylabel = "Prob >= $x$"
surv.plot()
decorate(xlabel=xlabel, ylabel=ylabel)
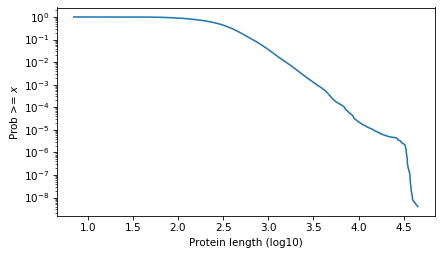
And here it is on a log scale.
surv.plot()
decorate(xlabel=xlabel, ylabel=ylabel, yscale='log')
plt.savefig("proteins1.png", dpi=300)
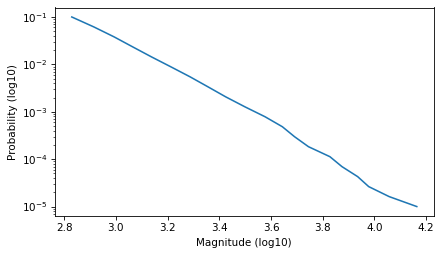
Fit a Power Law#
Select the part of the curve that looks like a straight line.
low, high = -1, -5
ps = np.logspace(low, high, 20)
qs = surv.inverse(ps)
plt.plot(qs, ps)
decorate(xlabel='Magnitude (log10)', ylabel='Probability (log10)', yscale='log')
And fit a line to it.
from scipy.stats import linregress
log_ps = np.log10(ps)
result = linregress(qs, log_ps)
result
LinregressResult(slope=-3.0982449194692387, intercept=7.8772243002229825, rvalue=-0.9986259969715762, pvalue=1.6492457998656753e-24, stderr=0.03832094735310883, intercept_stderr=0.13536901050675956)
Extend the line to the full domain.
log_ps_ext = result.intercept + result.slope * surv.qs
ps_ext = np.power(10, log_ps_ext)
surv_power_law = Surv(ps_ext, surv.qs)
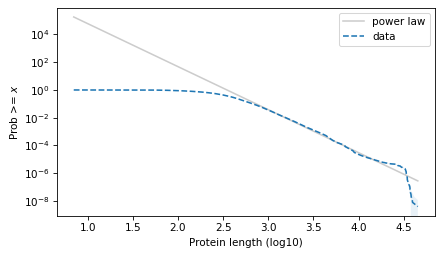
Here’s what it looks like on a log-log scale.
surv_power_law.plot(color='gray', alpha=0.4, label='power law')
plot_error_bounds(surv, count, color="C0")
surv.plot(ls='--', label="data")
decorate(xlabel=xlabel, ylabel=ylabel, yscale='log')
plt.savefig("proteins2.png", dpi=300)
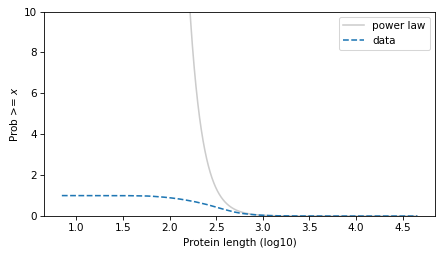
And on a linear y scale.
surv_power_law.plot(color='gray', alpha=0.4, label='power law')
surv.plot(ls='--', label='data')
decorate(xlabel=xlabel, ylabel=ylabel, ylim=[0, 10])
plt.savefig("proteins3.png", dpi=300)
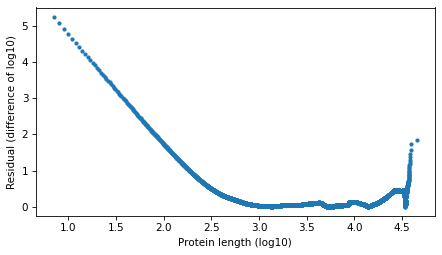
Compute residuals as differences in log survival function.
resid_power_law = np.log10(surv_power_law.ps) - np.log10(surv.ps)
plt.plot(surv.qs, np.abs(resid_power_law), '.')
decorate(xlabel=xlabel, ylabel='Residual (difference of log10)')
Fit a Student t#
First we’ll find the df parameter that best matches the tail, then the mu and sigma parameters that best match the percentiles.
from scipy.stats import t as t_dist
def truncated_t_sf(qs, df, mu, sigma):
ps = t_dist.sf(qs, df, mu, sigma)
surv_model = Surv(ps / ps[0], qs)
return surv_model
from scipy.optimize import least_squares
def fit_truncated_t(df, surv):
"""Given df, find the best values of mu and sigma."""
low, high = surv.qs.min(), surv.qs.max()
qs_model = np.linspace(low, high, 1000)
# match 20 percentiles from 1st to 99th
ps = np.linspace(0.01, 0.99, 20)
qs = surv.inverse(ps)
def error_func_t(params, df, surv):
mu, sigma = params
surv_model = truncated_t_sf(qs_model, df, mu, sigma)
error = surv(qs) - surv_model(qs)
return error
params = surv.mean(), surv.std()
res = least_squares(error_func_t, x0=params, args=(df, surv), xtol=1e-3)
assert res.success
return res.x
from scipy.optimize import minimize
def minimize_df(df0, surv, bounds=[(1, 1e6)], ps=None):
low, high = surv.qs.min(), surv.qs.max()
qs_model = np.linspace(low, high * 1.0, 2000)
if ps is None:
# fit the tail over these orders of magnitude
ps = np.logspace(-1, -5, 20)
qs = surv.inverse(ps)
def error_func_tail(params):
(df,) = params
mu, sigma = fit_truncated_t(df, surv)
surv_model = truncated_t_sf(qs_model, df, mu, sigma)
errors = np.log10(surv(qs)) - np.log10(surv_model(qs))
return np.sum(errors**2)
params = (df0,)
res = minimize(error_func_tail, x0=params, bounds=bounds, tol=1e-3, method="Powell")
assert res.success
return res.x
Search for the value of df that best matches the tail behavior.
df = minimize_df(5, surv, [(1, 100)])
df
array([22.58669028])
Given that value of df, find mu and sigma to best match percentiles.
mu, sigma = fit_truncated_t(df, surv)
df, mu, sigma
(array([22.58669028]), 2.4369128138364107, 0.3145746370844557)
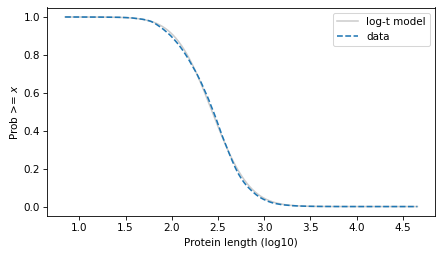
Evaluate the model over the range of the data.
surv_model = truncated_t_sf(surv.qs, df, mu, sigma)
surv_model.plot(color="gray", alpha=0.4, label="log-t model")
surv.plot(ls="--", label="data")
decorate(xlabel=xlabel, ylabel=ylabel)
plt.savefig("proteins4.png", dpi=300)
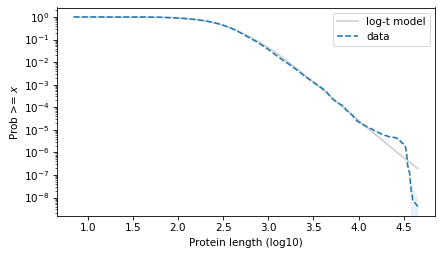
surv_model.plot(color="gray", alpha=0.4, label="log-t model")
surv.plot(ls="--", label="data")
plot_error_bounds(surv, count, color="C0")
decorate(xlabel=xlabel, ylabel=ylabel, yscale="log")
plt.savefig("proteins5.png", dpi=300)
resid_t = np.log10(surv_model.ps) - np.log10(surv.ps)
plt.plot(surv.qs, np.abs(resid_power_law), '.', ms=1)
plt.plot(surv.qs, np.abs(resid_t), '.', ms=1)
decorate(xlabel=xlabel, ylabel='Residual (difference of log10 SF)')
plt.savefig("proteins6.png", dpi=300)
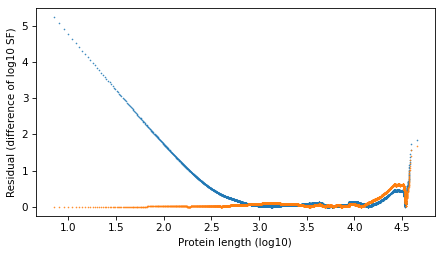
The results with the protein lengths are similar to the results with the function lengths.
The power law model is a little better in the extreme tail. They are about the same in the middle. And the t model is much better to the left of the median.
Allen Downey MIT License